Guidance for a Laboratory Data Mesh on AWS
Summary: This AWS Guidance accelerates the development of a scientific data management system that integrates laboratory data and software with cloud data governance, data discovery, and bioinformatics pipelines, while capturing important metadata along the way. Life sciences cloud engineering teams can use this Guidance to design systems to store, enrich with metadata, and search for scientific data sets. This can extend upon modern lab management systems, electronic lab notebooks (ELNs), and bioinformatics processing pipelines to create a scalable laboratory data mesh. This Guidance is primarily meant for cloud engineering teams who build solutions to support R&D, diagnostics, and manufacturing in life science organizations.
Introduction
Overview
This AWS Guidance accelerates the development of a scientific data management system that integrates laboratory data and software with cloud data governance, data discovery, and bioinformatics pipelines, while capturing important metadata along the way. Life sciences cloud engineering teams can use this Guidance to design systems to store, enrich with metadata, and search for scientific data sets. This can extend upon modern lab management systems, electronic lab notebooks (ELNs), and bioinformatics processing pipelines to create a scalable laboratory data mesh.
This Guidance is primarily meant for cloud engineering teams who build solutions to support R&D, diagnostics, and manufacturing in life science organizations.
Benefits
The architectures in this Guidance help achieve the following benefits:
- Creating operational efficiencies by linking lab systems such as electronic lab notebooks (ELNs) and lab information management systems (LIMS) to scalable, cloud data repositories
- Providing a flexible approach to cost-effectively store instrument data files in the cloud, across many laboratory domains
- Contextualizing the data stored in the cloud by continually updating metadata throughout its lifecycle
- Simplifying access controls to scientific data for users, across multiple enterprise domains
- Improving discoverability of historical data sets through keyword and semantic search. Pairing search with large language models (LLMs) for creating summarization and chat applications.
Digital Labs Strategy
This Guidance is closley related to several other AWS resources, which as a whole can help you to build a life sciences laboratory data mesh and bioinformatics platform.
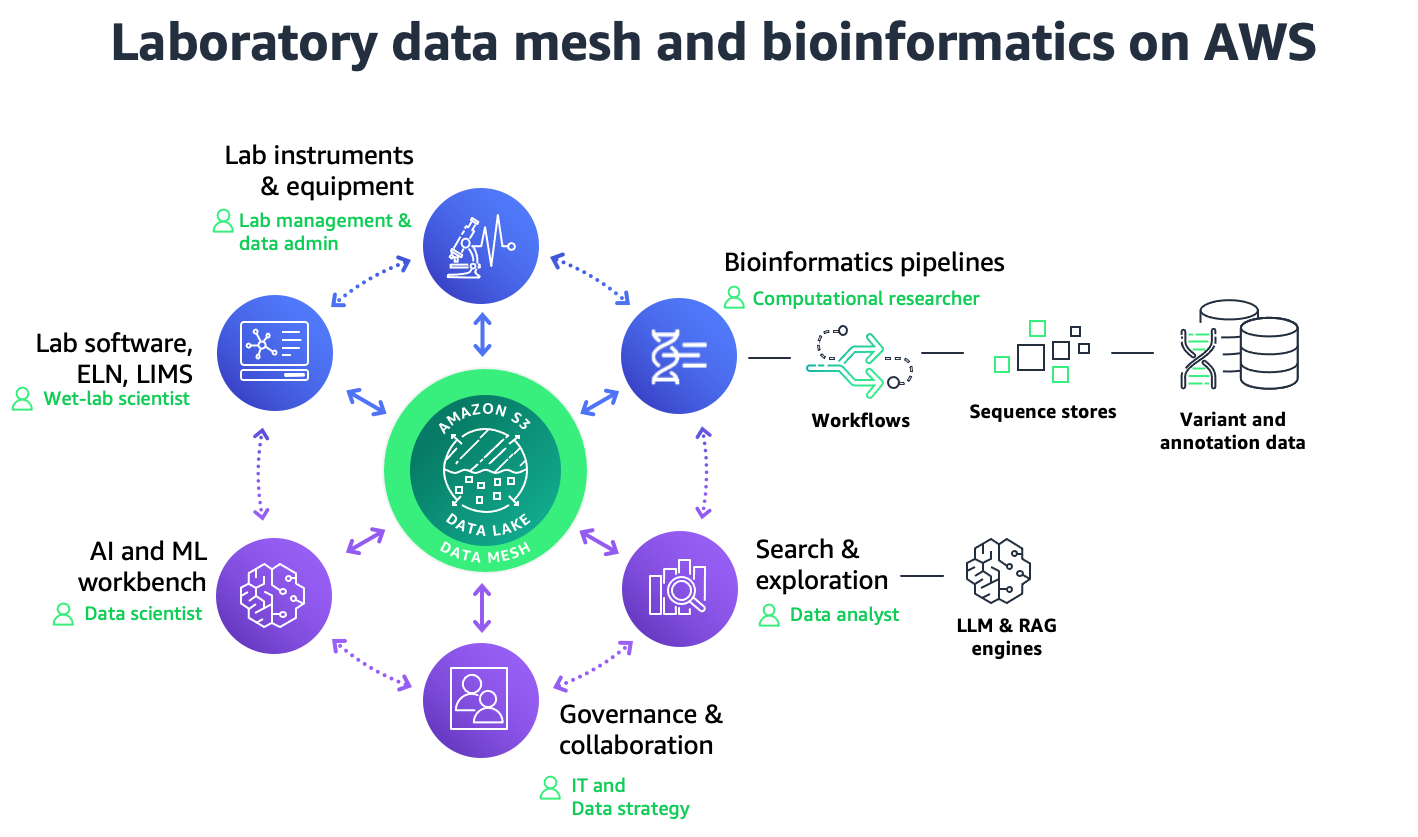
Figure 1: Laboratory data mesh and bioinformatics capabilities
In this Guidance, we go into detail on the implementation best practices for ELN/LIMS integration, metadata enrichment, and data discovery. This Guidance is a component of an overarching Digital Labs Strategy which includes other areas like lab event automation and machine learning.
In this Guidance, we go into detail on the interaction between cloud lab data management and cloud bioinformatics pipelines, specifically when it comes to capturing key metadata. For detail on how to build bioinformatics workloads on AWS, we refer to the Guidance for Development, Automation, Implementation, and Monitoring of Bioinformatics Workflows on AWS.
In this Guidance, we describe the use of AWS DataSync as the main service to automate the transfer of instrument data. For detail on how to augment DataSync with other scalability and monitoring features, we refer to the Life Sciences Data Transfer Architecture.
Concepts
- Data mesh principles: This Guidance lays a foundation for a self-service data platform where data consumers (scientists, bioinformaticians, business users) can discover, access, and use data while data producers (ELNs, LIMS, instruments) can build and maintain data products. The approach prioritizes the use of data lakes for the most effective and scalable storage for unstructured datasets like instrument data, while using purpose-built systems to provide the user interface and the industry-specific data models. A key principle is seamless data movement through event-driven architectures, to provide notifications between systems. Finally, we include unified policy management, centralized governance and audit, federated fine-grained access control and organization wide settings. More information on data mesh principles can be found at What is a Data Mesh?.
- Lab and Omics data: Unstructured and semi-structured data files are prevalent in research, development, biomanufacturing, and diagnostics organizations. This may include genetic sequence data, microscope images, microwell plate readouts, analytical spectra, and chromatograms. This also includes the processed outputs of algorithms, such as those in genomic secondary analysis. The data may be created by instruments coming from a variety of vendors.
- Metadata: Broadly, metadata is information that describes and explains data. Metadata may refer to 1) business metadata including study, project, laboratory protocol, and assay information, 2) operational metadata including algorithmic steps, compute tasks, and process outputs, 3) technical metadata like schema, format, and data types. The focus of this Guidance is on business (laboratory) metadata and operational metadata.
- Domain: A domain refers to a set of ELN/LIMS, experiment types, users, and instrument data types that have a certain set of metadata fields that are relevant to them. In this Guidance, we choose a example of a domain that uses metadata fields such as “group,” “site,” “platform,” “experiment_id,” and “sample_id.” This Guidance can be extended to multiple domains which may have different metadata fields.
- File-based data: Data that is stored and maintained in a file format is file-based data. Streaming data and Internet of Things (IoT) data are not covered in this Guidance but are covered in other Life Sciences Manufacturing and Supply Chain Solutions.
- Bioinformatics pipeline metadata: This Guidance enriches the output of bioinformatics pipelines with metadata that originates from the ELN/LIMS. This Guidance can be extended to capture more metadata pertaining to run information, to aid in downstream search.
Architecture overview
Design Principles
- The ELN or LIMS is the system of record for experiment metadata. Whenever relevant new data sets appears on the cloud (such as instrument data or bioinformatics outputs), the ELN/LIMS experiment record gets linked to that data.
- Amazon Simple Storage Service (Amazon S3) and AWS HealthOmics Sequence Stores are the cloud Data Stores for raw, processed, and final data sets, including active and archival. Therefore, whenever relevant experiment metadata appears in the ELN/LIMS, the cloud is updated with that metadata.
- An ELN/LIMS template defines the metadata fields that are in use for a domain. The cloud Metadata Store inherits those fields.
- Data sets should be discoverable through a search tool that is extensible across multiple labs, sites, divisions, and domains, secured under enterprise governance. Users should be able to find and request access to datasets based on keyword metadata search. This should be integrated with corporate identity and access management
- Data discovery should be extendable to LLM-based agents for conversational search or retrieval augmented generation (RAG).
System components
- ELN/LIMS on the laboratory’s network (may be on-premises or cloud based)
- Instrument data file server on the laboratory’s network
- Data Store on the cloud
- Metadata Store on the cloud
- Bioinformatics pipelines on the cloud
- Intelligent search that can extend to an LLM chat interface
Data flow
- User sets up an experiment in their ELN or LIMS which notifies the Metadata Store and Data Store of the experiment.
- When instrument data is collected at a future point in time, data moves to the cloud and is associated with the experiment metadata that was sent previously.
- Processing steps are captured. All new files are linked to the ELN/LIMS through the Metadata Store.
- Data is searchable in a discovery portal, and exploration can be extended into an LLM chat interface.
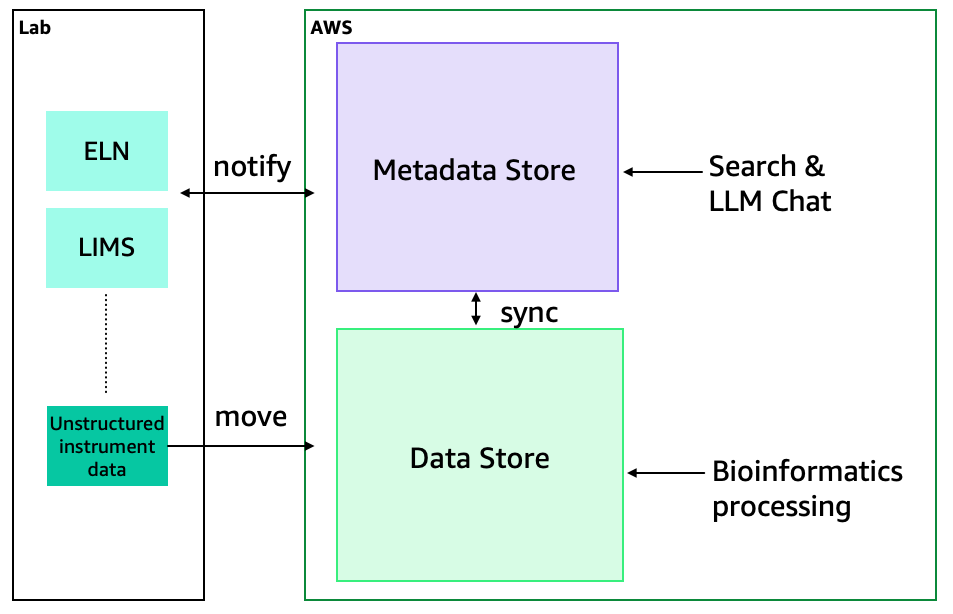
Figure 2: Design Principles. Whenever a user sets up an experiment in the ELN or LIMS, this notifies the Metadata Catalog and configures the Data Store to receive that experimental data. Later, when instrument data is collected, data moves to the Data Store that was pre-associated with the Metadata Store. Bioinformatics processing steps and output files are captured within the Data Store, and all new files are linked to the ELN/LIMS through the Metadata Store. Data is searchable and available through a chat interface.
Deployment options
This Guidance focuses on architectural design and implementation principles. When it comes to getting started, there are numerous ways to deploy the solutions, including building directly with AWS Services and adopting industry-ready AWS Partner Solutions.
- Cloud engineering teams who wish to build directly with AWS Services can use this guide to begin developing solutions immediately.
- For teams looking to use, or build on top of, industry-ready solutions we encourage engaging with AWS Life Sciences Partners who offer solutions that shorten the time it takes to deploy these architectures.
- For teams looking for hands-on support, AWS Professional Services and our global network of systems integration partners can provide consulting expertise.
Reference architecture
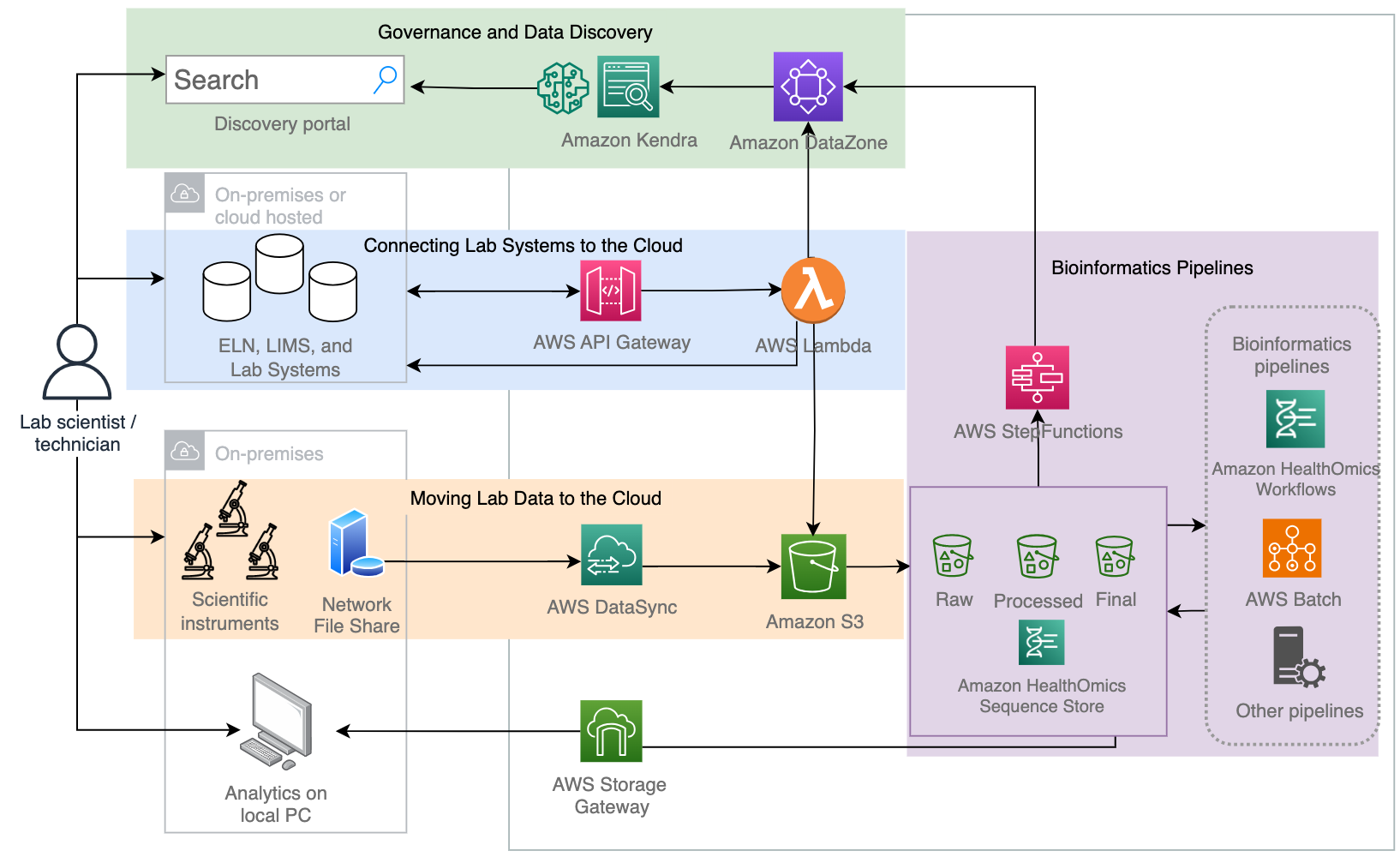
Figure 3: Architecture overview: I) Connecting Lab Software to the Cloud. When a scientist or technician sets up an experiment in the ELN or LIMS, this notifies the Metadata Catalog of that new experiment and configures the Data Store to receive that instrument data. II) Connecting Lab Instrument data to the Cloud. Later, when lab instrument data is collected, the data moves to the Data Store that was pre-associated with the Metadata Store. III). Connecting Bioinformatics Pipelines. Processing steps and output files are captured within the Data Store, and all new files are linked to the ELN/LIMS through the Metadata Store. IV.) Governance and Data Discovery. Data is governed and discoverable by metadata search, or by natural language search through a chat interface.
This Guidance is partitioned into 4 sections below:
- I. Connecting lab systems to the cloud
- II. Moving lab data to the cloud
- III. Bioinformatics pipelines
- IV. Governance and data discovery
Connecting lab systems to the cloud
Lab software like ELNs and LIMS need to have bidirectional communication with the cloud in order to 1) take advantage of scalable storage as an extension of the ELNs , 2) for the software to maintain the latest context about new files moved to or created on the cloud, which may reside outside of the ELN workflows, and 3) for cloud services to have contextual information about datasets, for improved data governance and enterprise search.
Initialization of ELN/LIMS
- Configure the ELN so that upon a Create Experiment action, Amazon API Gateway is called and sent the unique Experiment ID.
- Alternatively: If the ELN uses Amazon EventBridge natively, this can remove the need for using API Gateway. Instead, the system can be configure to create an EventBridge event.
Initialization of the Metadata Store
For every laboratory domain, defined by an ELN / LIMS template, an Amazon DataZone Project should be created.
- From the AWS Console, create an Amazon DataZone Domain. Enable the Blueprint for Default Data Lake.
- Within an Amazon DataZone portal, create a new Project. For example “RnD-Digital-Lab-Management”.
- Within the DataZone Project, create Business Glossaries to define the business metadata terms that are relevant for an organization. As an example, in this Guidance we use the following glossaries: group, source-region, site, and platform (for example, flow cytometry).
- Within the DataZone Project, create a Metadata Form to define the terms that are relevant for a particular ELN or LIMS template (for example, all of the metadata fields that are capture in a Revvity Signals ELN template for an Early Discovery team). These should be fields that represent the relevant experiment metadata that are helpful for searchability.
Organizations with large geographic diversity, multiple business units, and multiple lab domains will benefit from having well-planned business glossaries for organizing the company’s data.
- Use the Required checkbox for fields that are mandatory for an Experiment creation in the ELN.
- Example: In this Guidance we use the following metadata fields: (mandatory) system-domain, (mandatory) experiment_id, (mandatory) project_num, (mandatory) workflow, description, samples, related files.
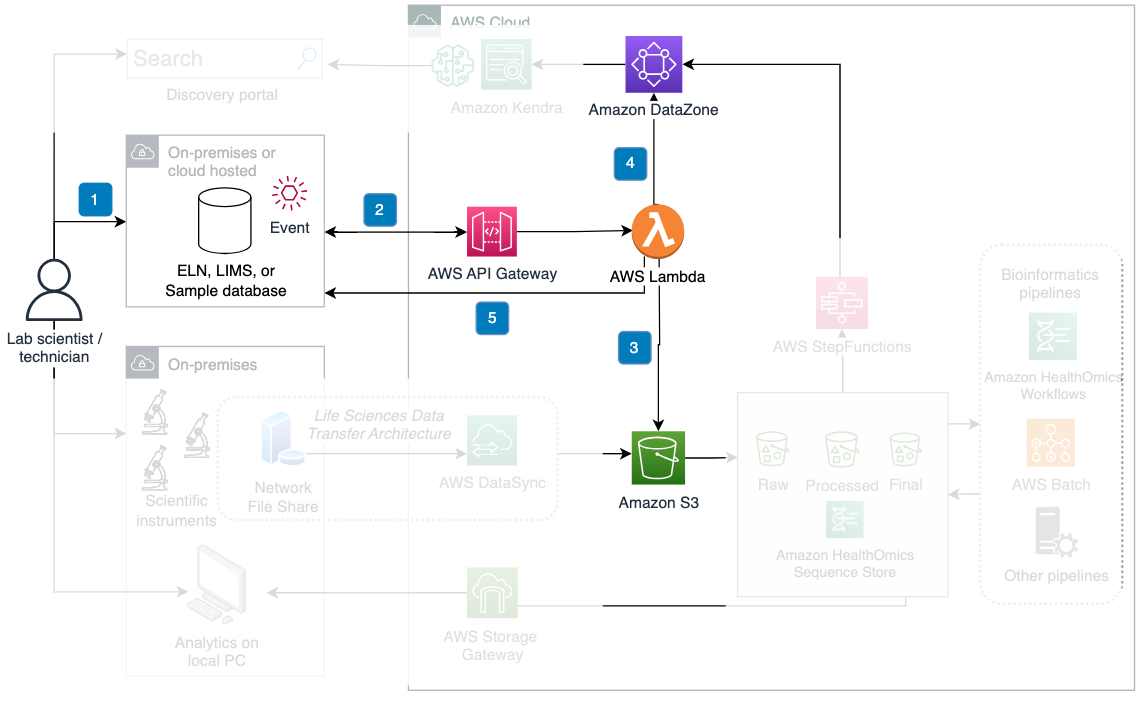
Figure 4: Data flow for connecting ELN/LIMS to the cloud.
Steps 1 and 2: Experiment creation and metadata retrieval
- A scientist sets up project or experiment metadata within the ELN, LIMS, or other experiment/testing database
- Upon the creation of an Experiment in the ELN, the ELN creates an event that calls an AWS API to send an Experiment ID. With the Experiment ID received, an AWS Lambda function calls an ELN’s API to retrieve all of the Experiment metadata that will be relevant in search.
Key considerations
- We assume the ELNs and LIMS to be the central user interface for the scientist to manage sample information, protocol information, assay information, and lab notes. Many modern ELNs and LIMS have event systems and an API that can be used to integrate with this Guidance.
- Assumption: The following require that the ELN/LIMS has an API for data retrieval requests, and has an event (bus) system which can generate events that call an external REST API.
- API Gateway uses a Lambda function and makes an API request back to the ELN, to retrieve more data for that Experiment ID. The selection of metadata to capture should be decided based on the most informative fields that will be helpful for associating with the instrument data, when performing search.
- Alternative: If the ELN uses EventBridge, this replaces the need for API Gateway. An EventBridge Rule should be used to launch the Lambda function. The Lambda makes a request back to the ELN as mentioned above.
- The selection of metadata to capture should align with the ELN/LIMS and the operating procedures of the lab. So, the ELN should be configured such that those metadata fields are mandatory fields before experiment creation.
An example of the experiment metadata that should be captured from the ELN is below:

Figure 5: Example of metadata collected from ELN.
Step 3. Setup of the Data Stores
- The Lambda sets up the Amazon S3 buckets as a scientific Data Store. The setup includes the naming of related folders, based on the unique Experiment ID. At this step, the Data Stores are empty. In addition to this, next-generation sequencing (NGS) sequence data can be stored in AWS HealthOmics.
Details For Amazon S3 Data Stores
- For a laboratory data mesh that will support diverse data types and instrument types, we recommend using Amazon S3 as the Data Store. It is a highly available, scalable cloud object storage service that stores data as objects within buckets.
- With the relevant Experiment Metadata obtained from the ELN, the Lambda function will set up an empty Data Store using that metadata in a hierarchy that is meaningful to that laboratory.
- We recommend a folder hierarchy similar to the one below. This is based on Defining S3 bucket and path names for data lake layers on the AWS Cloud and adapting it to the example laboratory metadata in the prior section. Blue boxes indicate prefix names that will be populated based on the metadata obtained from the ELN. Note: /source-system will not be populated at this time.
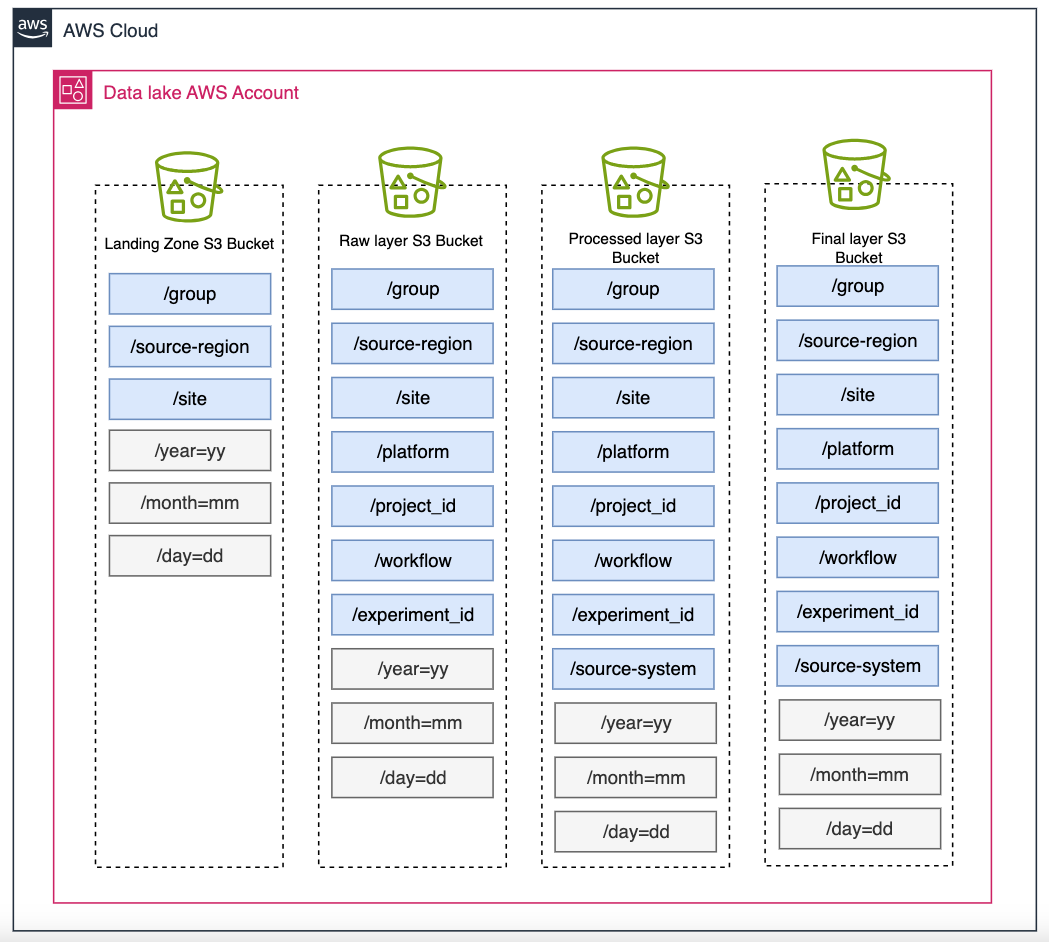
Figure 6: Bucket organization.
Details For AWS HealthOmics Data Stores
- For a laboratory data mesh with the primary use case of genomics and transcriptomics, we recommend using HealthOmics as the Data Store
- One Time Setup: A HealthOmics Reference Store and Sequence Store need to be set up once, prior to the first experiment being created, and can be used for all subsequent experiments. At this point it is empty.
Steps 4 and 5. Writing to the Metadata Catalog and linking back to ELN/LIMS
- The Lambda function writes the metadata that it has collected into an Amazon DataZone Metadata Catalog. This is done by creating DataZone Data Assets, adding Metadata Forms to those assets, and assigning the Metadata to those fields.
- Lambda calls the ELN’s API to add the location of the new Data Asset to the Experiment entity within the ELN.
Introduction to data assets in Amazon DataZone Data Assets are an anchor point for a collection of files in S3. The location of the Data Asset can be sent to the ELN/LIMS as a location where all data lives pertaining to that experiment. Experiment Metadata is added to a Data Asset. Access is granted based on the Data Asset.
Similar to how the data store was set up with three buckets-for raw, processed, and final files–so do the Data Assets need to be set up with three versions. For each experiment, there should be three (3) Data Assets set up: one for raw, one for processed, and one for final.
Details
- Create Data Assets using the API function CreateAsset().
- Recommendations:
- For Name, use “DataAsset-“ followed by the Experiment_ID, followed by ”-raw”, “-processed”, and “-final”.
- For the S3 location ARN (known in the API as externalIdentifier), use the location set up for the experiment_ID folder created in S3, for the raw, processed and final buckets.
- As an example:
- Name: DataAsset-experiment_flow_Brd9_CAS5_knockdown_16_2-RAW
- S3 location ARN: arn:aws:s3:::[bucket_name]-rawbucket/group_earlydiscovery/source-region_cambridge/site_building032/platform_flow/project_123456/workflow_protocol_7xb-flow/experiment_flow_Brd9_CAS5_knockdown_15/
- This example should be repeated for processed and final as well.
- For the three (3) data Data Assets that were created, the Lambda function adds metadata to them. To do this, use the formsInput parameter to add the Metadata Form (created in Metadata Store Initialization section), and populate the Metadata Form fields with the metadata that was retrieved from the ELN API.
- The Lambda function publishes the three Data Assets using CreateListingChangeSet().
- Finally, the Lambda should notify the ELN of the location of those Data Assets. To do this, Lambda writes the location of the three Data Assets to the ELN Experiment using the ELN’s API.
Example:
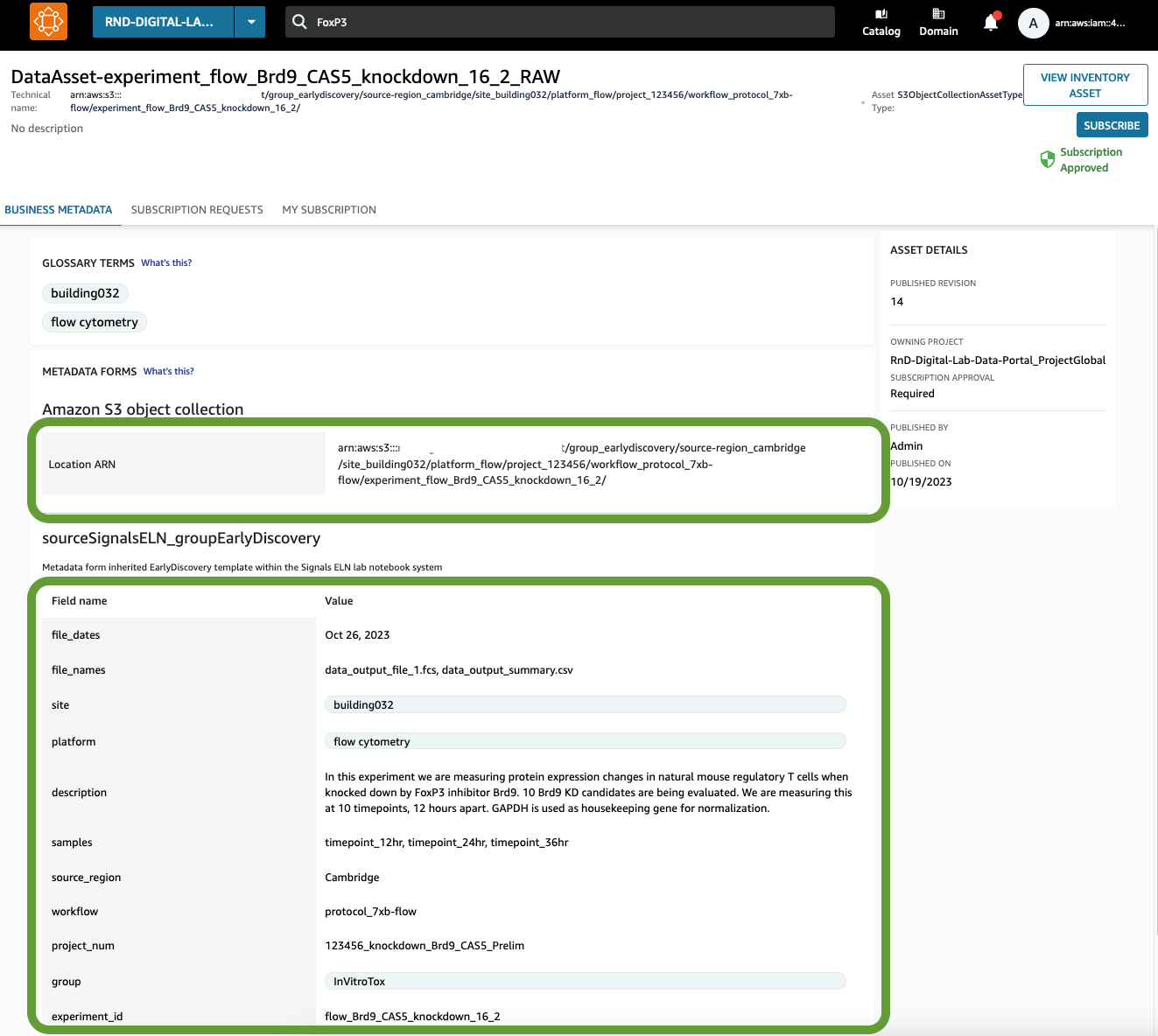
Figure 7: Here is a Data Asset for Experiment ID “flow_Brd9_CAS5_knockdown_16_2”, for the raw data. There are two aspects of a Data Asset: the physical location of the data (pointing to the raw S3 bucket), and the metadata that was populated by the ELN via the Lambda.
What if the ELN metadata changes in the future? • It is anticipated that scientists may update or add new metadata to the ELN over time, such as notes, conclusions, or corrections. The ELN should be configured so that when the experiment is updated (for example, through a Change Order Signing action) then the API Gateway is called again. • Then, when the Lambda retrieves the updated ELN information, Lambda should use CreateAssetRevision to update metadata through the formsInput parameter. • The Lambda function should re-publish the Data Assets using CreateListingChangeSet().
Moving lab data to the cloud
Lab instrument data can be automatically moved to the cloud to make it accessible to elastic cloud computing for data discoverability and for cost savings. A component of this is found in the blog How to move and store your genomics sequencing data with AWS DataSync.
Figure 7: Transferring lab instrument data to the cloud.
Step 6. Run lab instruments to collect lab data.
- Scientists run instruments to collect data and save data to a network-accessible folder being monitored by AWS DataSync.
Details
- The scientist or technician should initiate a run and save the data to a network-accessible file share, such as redundant arrays of independent disks (RAID)-enabled storage area network (SAN) or network-attached storage (NAS).
- The instrument or instrument control software must save the output files so that the Experiment ID is included in one of two ways:
- A) Experiment ID is added to the end of the filename
- B) Experiment ID is added to the end of the folder name that contains the run outputs.
- Note: Option B can be useful when an instrument run outputs multiple files such as with NGS
- If the instrument run is operated by a run plan that is generated by the ELN or LIMS, the output filenames or folder names can be configured when that run file is made.
- If the instrument run plan is set up manually by the scientist or technician, that user will need to ensure the instrument files are outputted with either A or B satisfied.
Step 7. Ingest lab data to Data Store
- AWS DataSync ingests the data into the Landing Zone bucket, within the Data Store.
Considerations
- AWS DataSync can be used for the performant transfer of up to terabyte size files and operates on a schedule or an event-driven basis. Briefly, a DataSync Agent is deployed on the laboratory network, the source location specifies where scientific data is being generated, and the target location specifies the S3 Landing Zone bucket as the location to move the data to.
- A standard DataSync deployment is described in the Getting started with AWS DataSync.
- Labs with high volume data movement or 24x7 operations may need additional monitoring and scheduling features. This is described in the blog How to move and store your genomics sequencing data with AWS DataSync.
Steps 8 and 9. Move the data into the Data Store and update Metadata Catalog and ELN
- The writing of the data to the Data Store invokes an event, initiating AWS Step Functions. The StepFunction will import the instrument data from the Landing Zone into the relevant Raw bucket within Amazon S3, and/or into the relevant Read Set within the AWS HealthOmics Sequence Store.
- The StepFunction adds the file names, creation date, and other metadata that is extracted from the files to the Metadata Store for the Raw Data Assets of the relevant Experiment.
Details For S3 Data Stores
- When data appears in the S3 Landing Zone bucket, an S3 event will invoke a Lambda function that will move the data from the Landing Zone Bucket to the Raw Bucket in the relevant Experiment ID folder. (Per Step 5, the relevant Experiment ID will be in the suffix of the filename or the folder name).
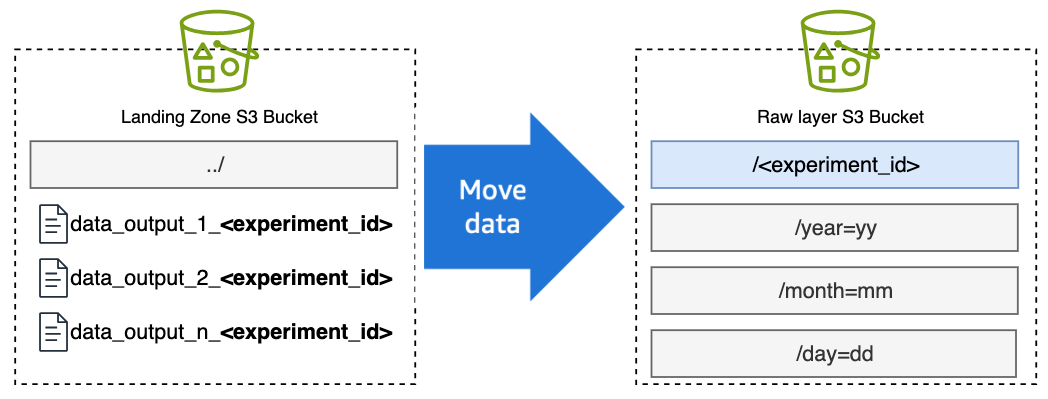
Figure 8: Transfer of instrument data to the cloud via DataSync.
- Using S3 bucket events, invoke a StepFunctions state machine, which is used to capture metadata and write it to the relevant DataZone Data Asset.
- The StepFunction should append the name of the file to the file_names metadata field for the DataAsset for the relevant Experiment ID.
- The StepFunction should append the creation date of the file to the file_dates metadata field for the Data Asset for the relevant Experiment ID.
Optionally, at this step the StepFunction may also be used to extract additional metadata from the contents of the data file, such as headers, config files, or summary files. That metadata should then be added to the Data Asset.
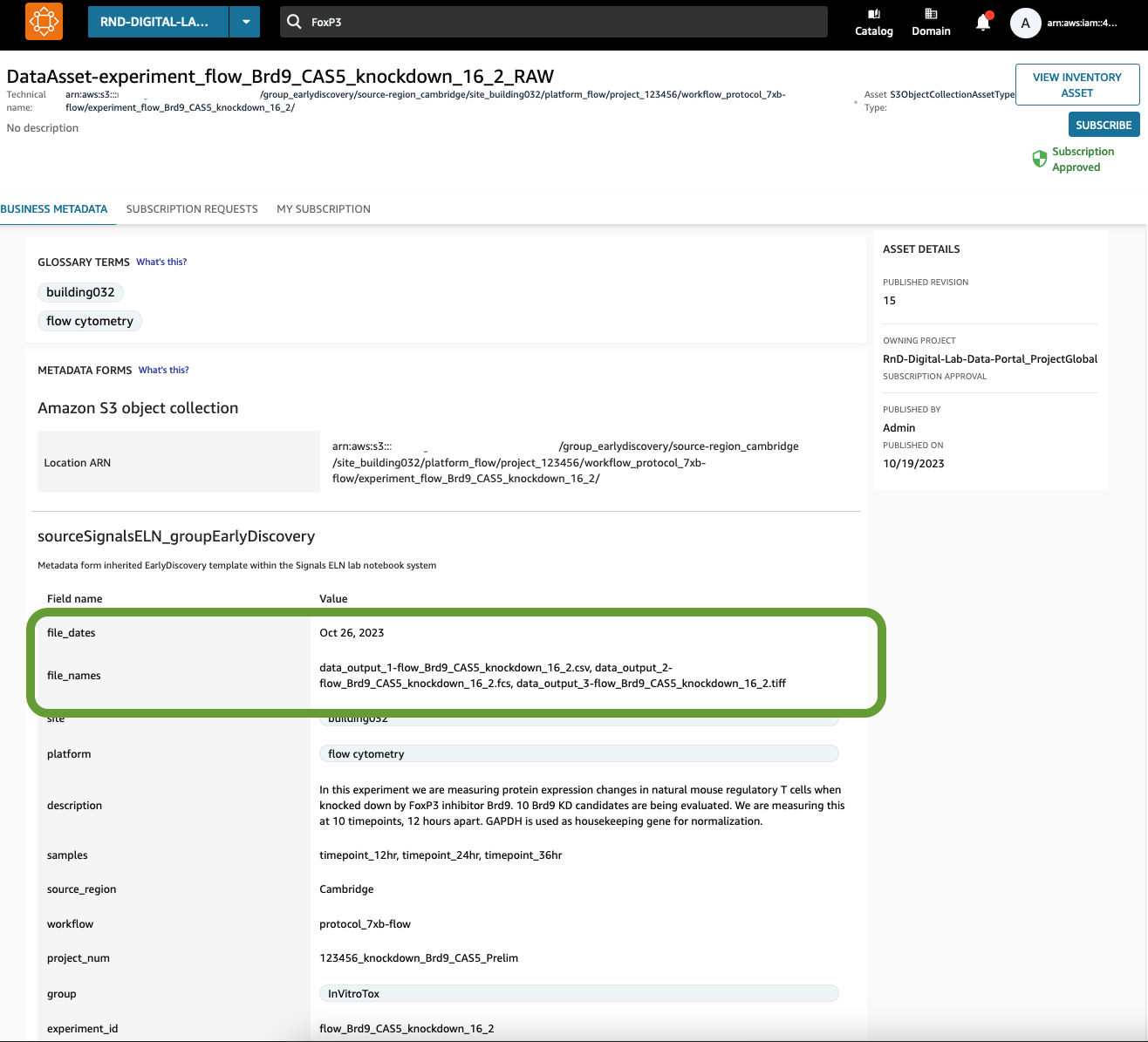
Figure 9: The names of the files and the date of their creation is appended into the metadata fields in the Data Asset. This will allow users to search for these assets using metadata terms within the file names.
Details For HealthOmics Data Stores
- When data appears in the S3 Landing Zone bucket, an S3 event will invoke a StepFunction.
- The StepFunction will import the instrument files (including FASTQ, BAM or CRAM files) into Read Sets within a HealthOmics Sequence Store. Assumption: One Sequence Store has been set up which will serve this ELN instance or lab.
- The StepFunction will use the Experiment ID of the filename or folder name, to retrieve experiment metadata from the Metadata Store (such as sample_id, project_id, workflow, flowcell_id, sample_id).
- The StepFunction will add Read Set Tags to the Read Set that was created, using the metadata that was retrieved from the Metadata Store
- The StepFunction will receive the Read Set URI which was created
- The Step Function will write the Read Set URI to the Metadata Store, under the metadata field readsets.
Bioinformatics pipelines
At this point, instrument data will be located in the raw bucket, and the Metadata Store will have contextual information about that data. Here we discuss how to submit that data to bioinformatics or high performance compute (HPC) processing in a way that maintains association with the Metadata Store.
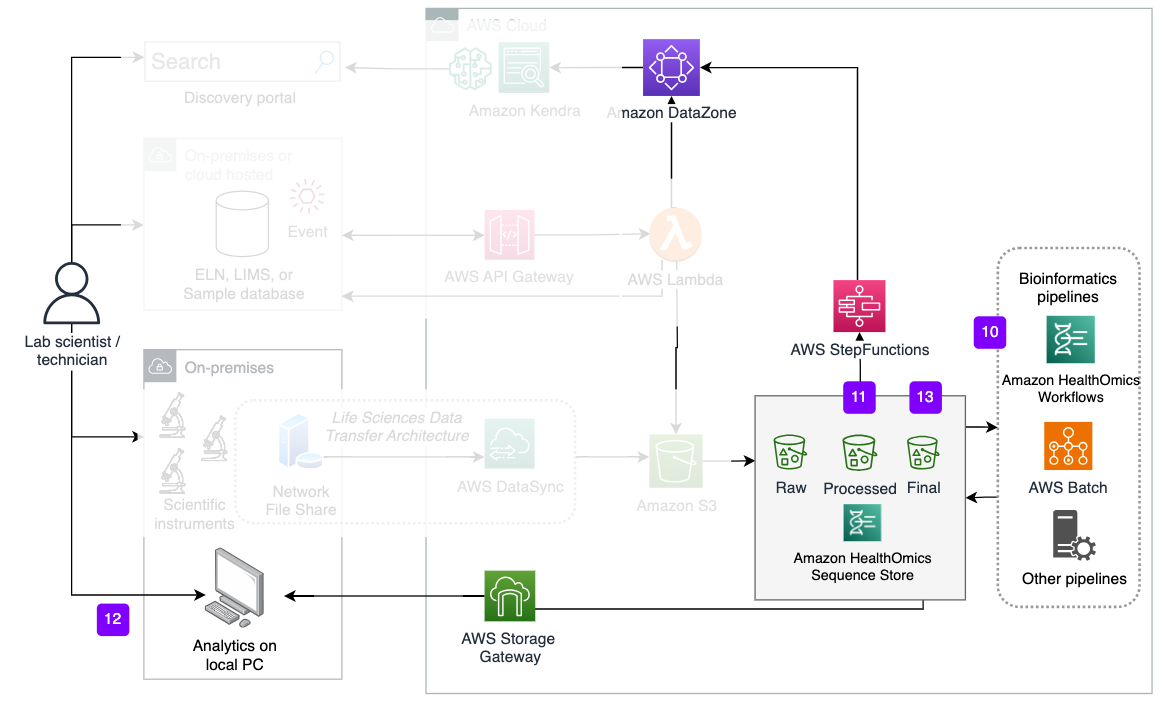
Figure 10: Launching bioinformatics pipelines.
Steps 10-13. Data processing and capturing provenance of outputs
- An S3 bucket event initiates a bioinformatics pipeline run using the Raw bucket as the data source. Bioinformatics output files are written to Processed bucket.
- The StepFunction adds the file names, creation date, and other metadata that is extracted from the files, to the Metadata Store for the Processed Data Assets of the relevant Experiment.
- Optionally, AWS Storage Gateway mounts onto the local network for users to access the Processed bucket for local analysis or report generation
- Locally-generated files are written to the Final bucket. The StepFunction posts the name and date to the Processed Data Asset in the Metadata Store
Details for cloud bioinformatics pipelines
- Once files appear in the Raw bucket under the relevant Experiment ID, a Lambda function starts a bioinformatics run using AWS HealthOmics Workflows, AWS Batch, or other system.
- An AWS HealthOmics or Batch workflow is launched with the input location set as a folder of the relevant experiment ID in the S3 Raw bucket.
- After successful completion of the workflow, the outputs are written to the S3 Processed bucket, under the relevant experiment ID, in a subfolder labeled by the name of the system. For example a workflow system could be /awshealthomics, /awsbatch, or another system.
- The appearance of the files in the output location will invoke a StepFunction.
- The StepFunction will post to the Metadata Store to add the name of the files and the creation date, to the Processed Data Asset of the relevant Experiment
Optionally, at this step the StepFunction may also be used to extract additional metadata from the contents of the bioinformatics output files, such as headers, config files, or summary files. That metadata should then be added to the Data Asset.
Details for local bioinformatics tools
- When local processing is needed, such as the use of GraphPad, PowerPoint, or other lab software, Storage Gateway should be used as the user’s access-point to the available data.
- Users should be given read-only access to the Raw and Processed buckets, and write access to the Final bucket. The Final bucket is where manually saved files like PDFs, PowerPoints, and other local processing outputs should be saved.
- Then, when output files appear in the Processed Bucket, notify the StepFunction of the appearance of the new file.
- The StepFunction should post to the Metadata Store to add the name of the files and the date, to the Final Data Asset of the relevant Experiment
Governance and data discovery
By this step, instrument data has been collected and associated with laboratory metadata. The lab metadata now provides rich context for the associated data sets and can be used to power a search tool, for lab users to discover, access and use the data. Large language models (LLMs) can also be used in combination with semantic search, to create conversational data exploration tools (chatbots).
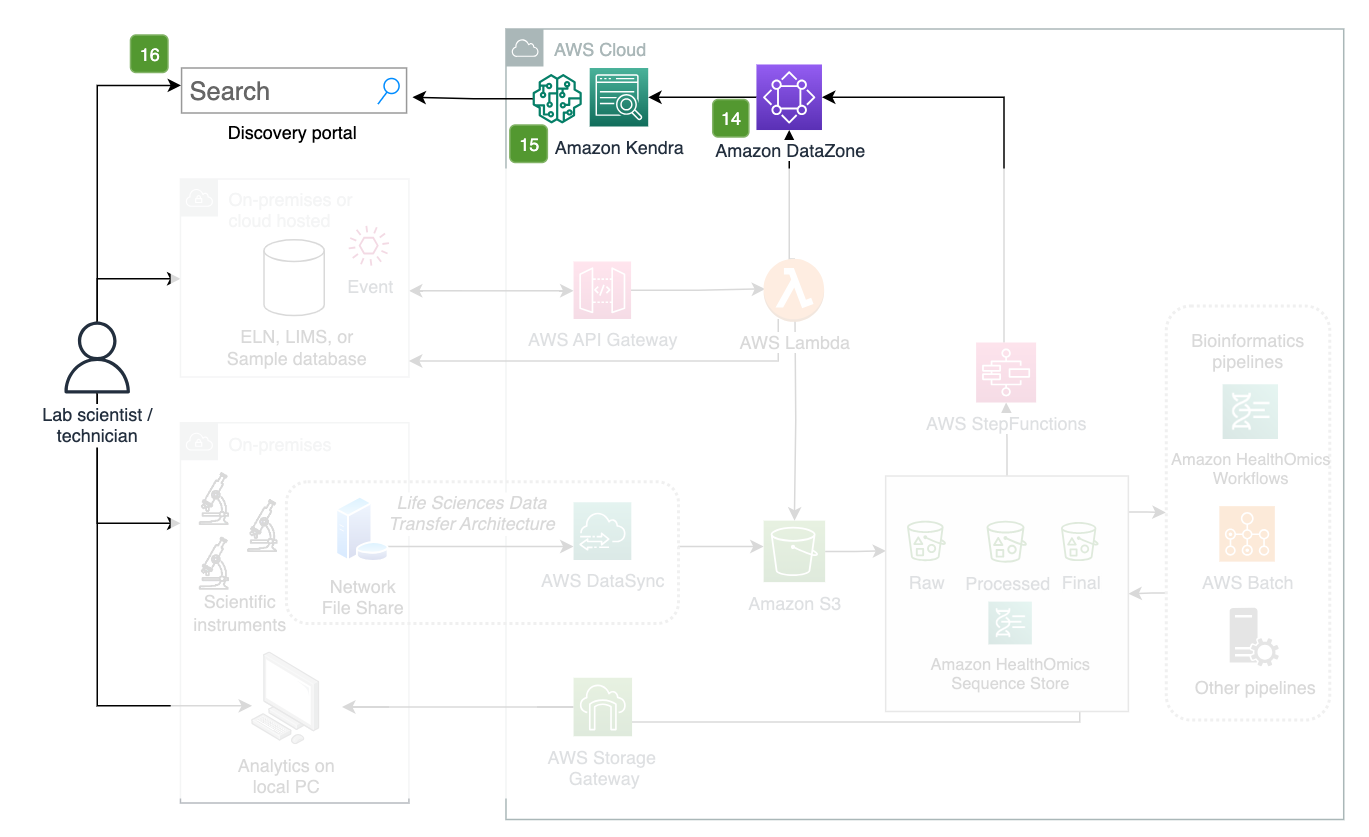
Figure 11: Governance and discovery portal.
Steps 14 - 16. Discovery portal
- Amazon DataZone is a tool for searching for files, requesting access, and granting access
- Amazon Kendra can be used to index the contents of data files and can be paired with an LLM for data summarization.
- Data and metadata can be semantically searched by a user to discover datasets, gain access to, and analyze datasets.
Amazon DataZone Portal
Amazon DataZone is a set of APIs and a data management portal to discover, analyze, and report on data. In the DataZone Portal, scientists and business users can search for Data Assets using keywords that are found in the metadata that originated from the ELN. For example, these may be searches for sample id, experiment id, group, platform, file names, dates, or keywords within the experimental description. These searches will return a list of Data Assets that have association with those keywords, which are collections of S3 objects.
To gain access to the Data Asset, users select the “Subscribe” button in the DataZone portal. If the user is a member of the Project, approval is immediate. Otherwise, an approval workflow reaches the Data Steward in the DataZone Project to approve the subscription request.
For teams that wish to use their own discovery portal for search and access controls, they can use the DataZone API to embed search, subscribe, and approval functionality within their application.
Once the subscription request is approved, DataZone sends an event to Amazon EventBridge. A pre-configured EventBridge rule triggers a Lambda function that is responsible for reading information from the event and granting permission to the identity trying to get access to the files within the Data Asset.
From this step, users can use multiple routes to read the data, process the data, and gain insights. For example, these files may be processed by bioinformatics pipelines on the cloud, outlined in Guidance for Multi-Modal Data Analysis with AWS Health and ML Services. Other examples include How Thermo Fisher Scientific Accelerated Cryo-EM using AWS ParallelCluster and Large scale AI in digital pathology without the heavy liftin). The files may be sent to SageMaker for machine learning predictions or for training, or they may be downloaded for local viewing and analysis.
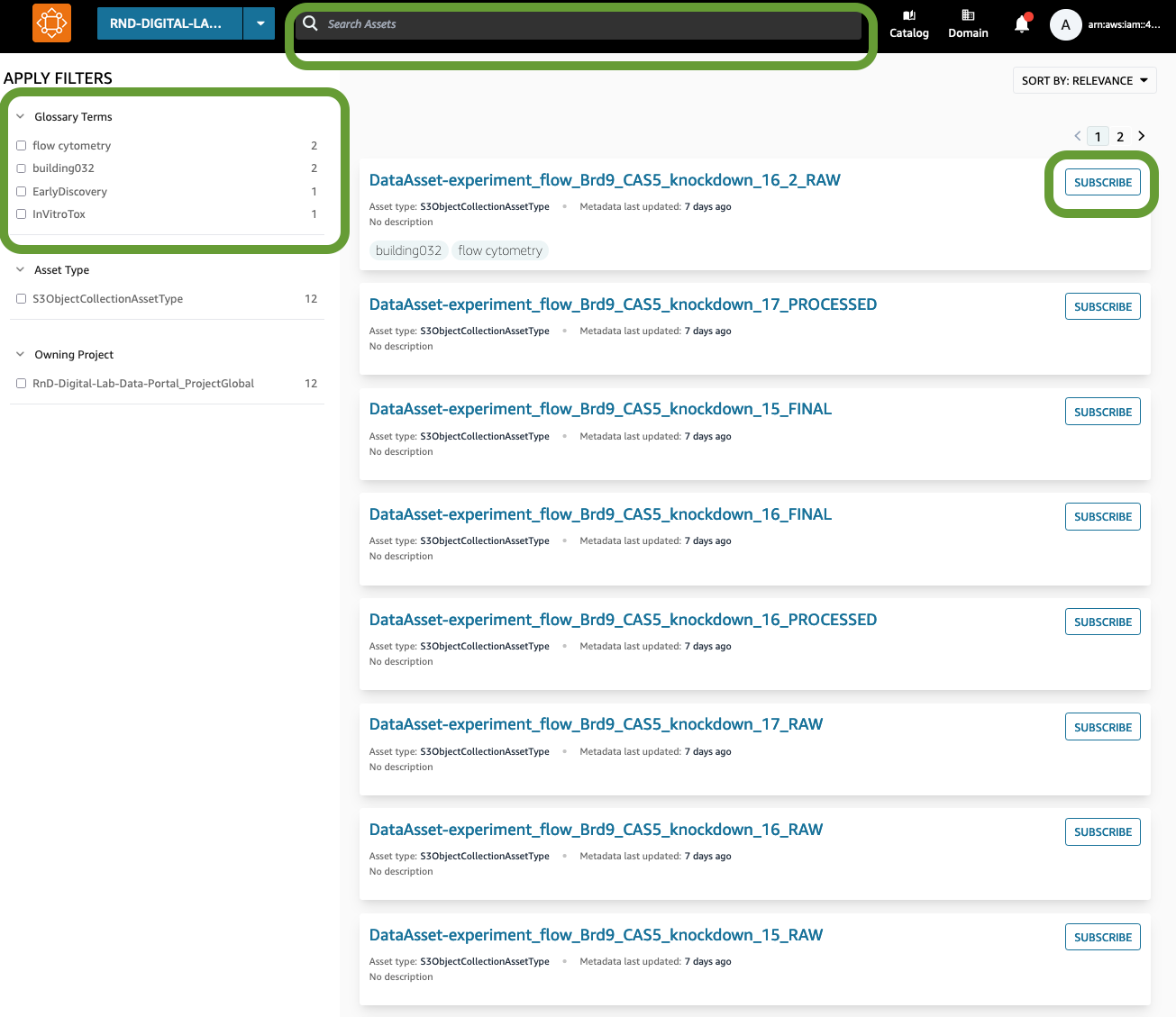
Figure 12: The DataZone Data Portal allows users to search by metadata to find Data Assets, filter by glossary terms, and subscribe to gain access. Users who are members of a Project will be granted access automatically while non-members will wait for a data steward to grant the access.
Custom Portal Applications with Kendra
For some data types, discoverability can be improved by not only using the laboratory metadata, but also by searching through the contents of the data sets. Some cases may include datasets that contain a corpus of text, datasets that have instrument-specific metadata embedded, or files that have other identifying keywords in their contents. To index the contents of the data files, Amazon Kendra is a managed service for indexing data and conducting semantic search. For teams that wish to use their own discovery portal for search, they can embed the Kendra API as the search engine within in their application.
Amazon Kendra APIs may be used within a custom Discovery Portal application that your organization creates, to enable this search. In combination with Kendra, large language models (LLMs) can be used to generate summaries of search results, and also create conversational experiences. Discover more examples to Quickly build high-accuracy Generative AI applications on enterprise data using Amazon Kendra, LangChain, and large language models and Interact Conversationally with AWS HealthLake.
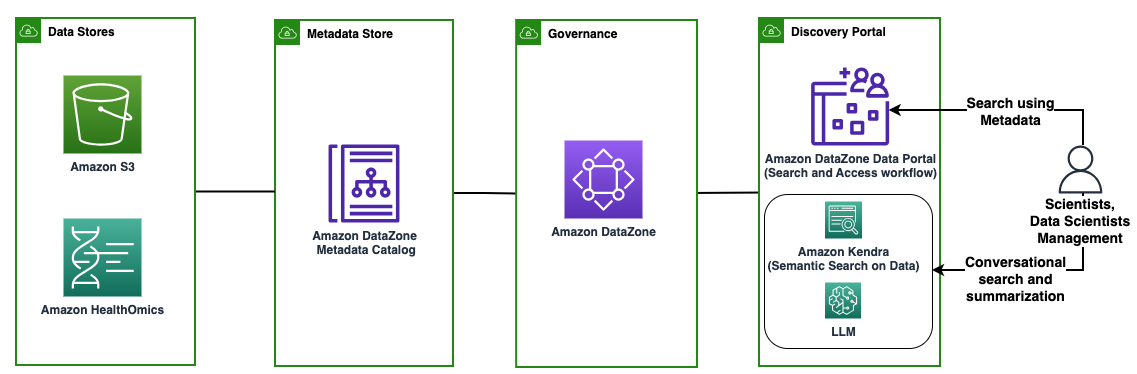
Figure 12: Summary of data productization approach. From left to right, this Guidance shows how to use data stored in physical Data Stores, turn them into data products through the Metadata Store, govern the access to them, and make them discoverable through a metadata search portal or through natural language search.
Conclusion
This Guidance helps create a laboratory data mesh composed of instrument data collection, lab metadata enrichment, data processing, and discoverability. With this architecture, cloud engineering teams can help life science organizations generate greater value from their data, streamline the use of their data, increase productivity through cloud elasticity, and accelerate the time to developing insights from laboratories. To learn more about what AWS can do for laboratory data mesh development, contact your AWS Respresentative.
Authors
- Lee Tessler - Principal, Technology Strategy, Healthcare & Life Sciences
- Yegor Tokmakov - Senior Solutions Architect, Healthcare & Life Sciences
- Beril Uenay - Associate Solutions Architect, Healthcare & Life Sciences
- Nadeem Bulsara - Principal Solutions Architect, Genomics
- Sujaya Srinivasan – Global Public Sector Genomics Tech Lead
- Subrat Das - Senior Solutions Architect, Healthcare & Life Sciences
- Navneet Srivastava - Principal, Data Strategy, Healthcare & Life Sciences
Notices
Customers are responsible for making their own independent assessment of the information in this document. This document: (a) is for informational purposes only, (b) represents AWS current product offerings and practices, which are subject to change without notice, and (c) does not create any commitments or assurances from AWS and its affiliates, suppliers or licensors. AWS products or services are provided “as is” without warranties, representations, or conditions of any kind, whether express or implied. AWS responsibilities and liabilities to its customers are controlled by AWS agreements, and this document is not part of, nor does it modify, any agreement between AWS and its customers.